
Contact information
Research groups
Colleges
David Grainger
BMedSc (Hons), DPhil
Postdoctoral Fellow in Developmental Haematopoiesis
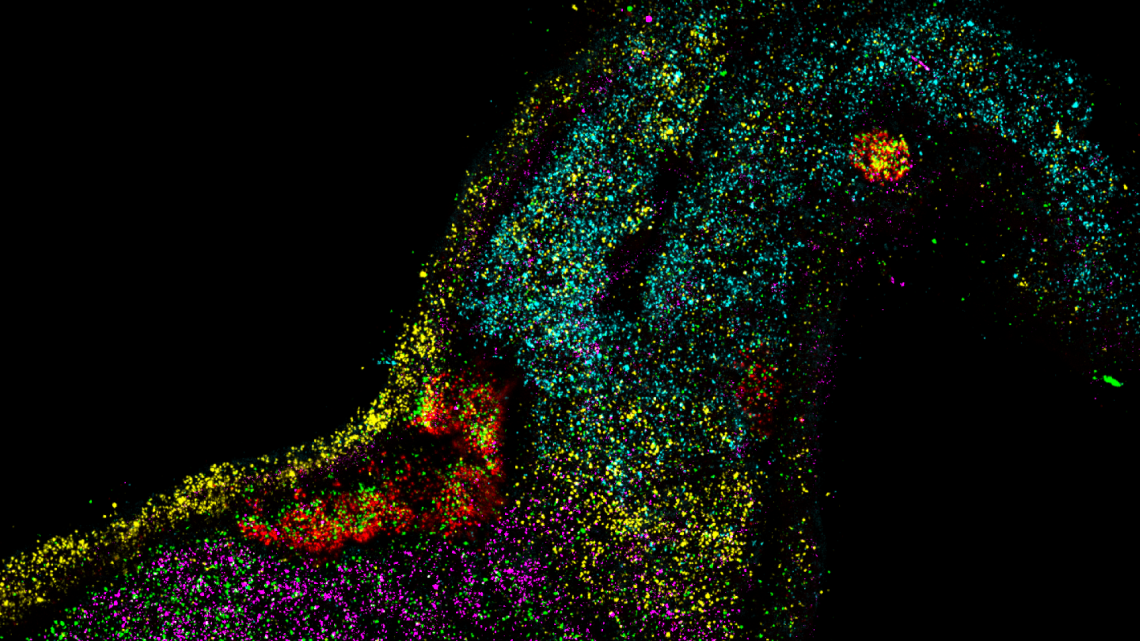
Spatial and Molecular Characterisation of the Cellular Niches Interacting with Intra-Embryonic Haemato-Endothelial Progenitors
I completed my undergraduate degree in Medical Sciences at the University of Birmingham in the summer of 2017. I moved to the University of Oxford in October 2017 to join Professor Catherine Porcher’s laboratory in the Weatherall Institute of Molecular Medicine to study for my DPhil in Medical Science.
My research interests include tissue-level imaging, lineage tracing and the diversification of cell lineages with a current focus on haematoendothelial lineages. My DPhil project aims to better characterise the specification of blood fated mesoderm in the mouse embryo in order to improve in vitro differentiation of human embryonic stem cells into haematopoietic lineages. This project utilises novel cre-based fate mapping models in combination with multiplexed single-molecule imaging and single-cell RNA-seq methods.
Media

Publications
David E. Grainger*, Vivien W. Ho*, Hedia Chagraoui, Catherine Porcher,
Specification of the haematopoietic stem cell lineage: From blood-fated mesodermal angioblasts to haemogenic endothelium, Seminars in Cell & Developmental Biology, 2022, ISSN 1084-9521, https://doi.org/10.1016/j.semcdb.2022.01.008. * denotes equal contribution.
Volpe, G., D. S. Walton, D. E. Grainger, C. Ward, P. Cauchy, D. Blakemore, D. J. L. Coleman, P. N. Cockerill, P. Garcia and J. Frampton (2017). "Prognostic significance of high GFI1 expression in AML of normal karyotype and its association with a FLT3-ITD signature." Scientific Reports 7(1): 11148


